Ensemble regression and calibration-in-time
Nick McKay
2025-05-31
Source:vignettes/regression.Rmd
regression.Rmd- Introduction to geoChronR
- Age-uncertain correlation
- Age-uncertain regression and calibration-in-time
- Age-uncertain spectral analysis
- Age-uncertain PCA analysis
Ensemble Regression and Calibration-in-time
Here, we replicate the analysis of Boldt et al. (2015), performing age-uncertain calibration-in-time on a chlorophyll reflectance record from northern Alaska, using geoChronR.
The challenge of age-uncertain calibration-in-time is that age uncertainty affects both the calibration model (the relation between the proxy data and instrumental data) and the reconstruction (the timing of events in the reconstruction). geoChronR simplifies handling these issues.
Let’s start by loading the packages we’ll need.
library(lipdR) #to read and write LiPD files
library(geoChronR) #of course
library(readr) #to load in the instrumental data we need
library(ggplot2) #for plottingLoad the LiPD file
OK, we’ll begin by loading in the Kurupa Lake record from Boldt et al., 2015. We’ll download the file from lipdverse, (using purrr::insistently to avoid issues with buildling) but you could also load it from a local file.
readLipdInsistently <- purrr::insistently(f = lipdR::readLipd,quiet = TRUE)
K <- readLipdInsistently("http://lipdverse.org/geoChronR-examples/Kurupa.Boldt.2015.lpd")## [1] "Loading 1 datasets from /var/folders/tp/bfmjfn9s0hd59bm9z80j3mgm0000gn/T//RtmpXsMOOQ/Kurupa.Boldt.2015.lpd..."
## [1] "reading: Kurupa.Boldt.2015.lpd"Check out the contents
sp <- plotSummary(K,paleo.data.var = "RABD",summary.font.size = 6)## [1] "Found it! Moving on..."
## [1] "Found it! Moving on..."
## [1] "Found it! Moving on..."
## [1] "Found it! Moving on..."
## [1] "Found it! Moving on..."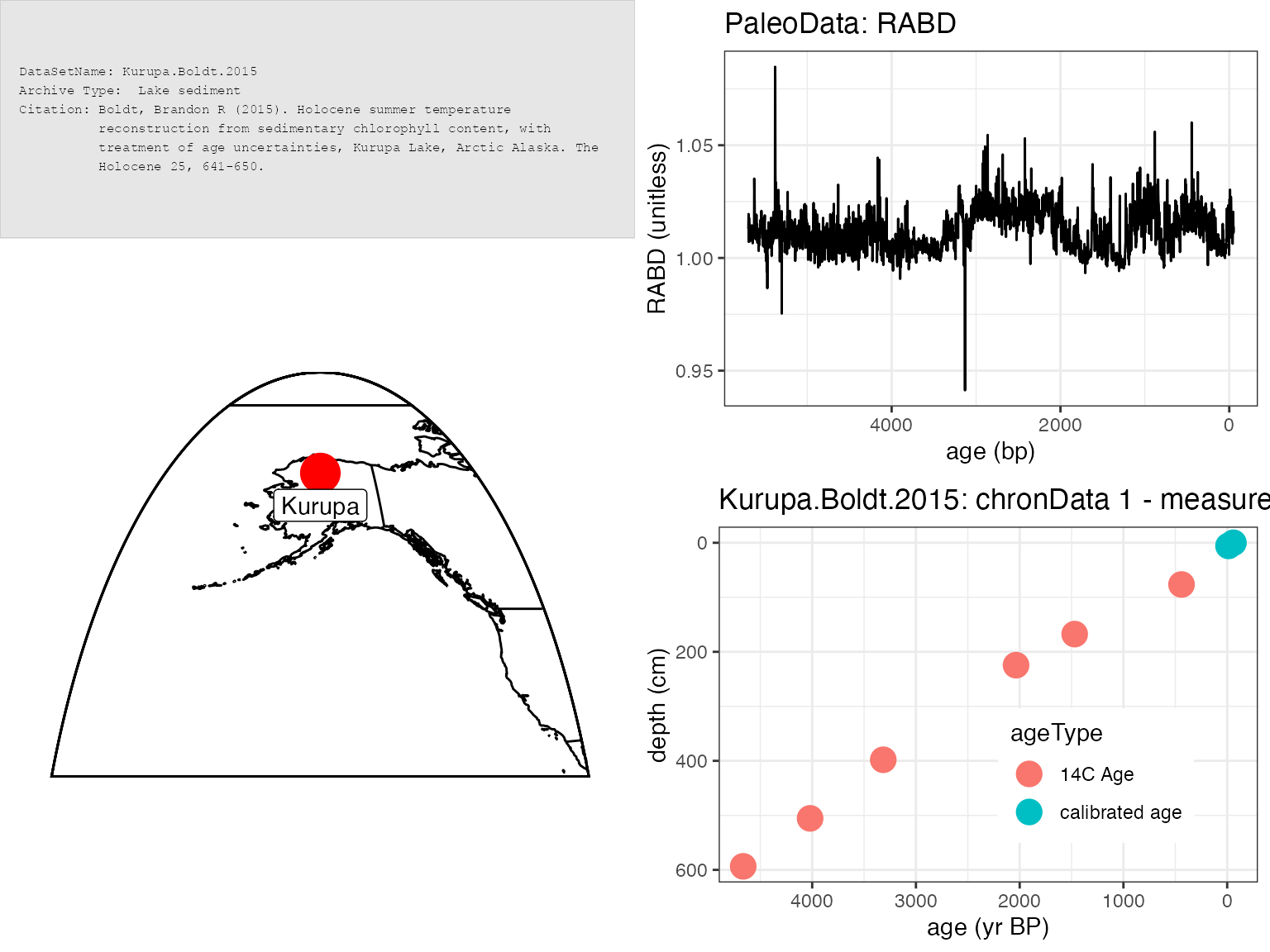
print(sp)## TableGrob (4 x 4) "arrange": 4 grobs
## z cells name grob
## 1 1 (1-1,1-2) arrange gTree[GRID.gTree.11]
## 2 2 (1-2,3-4) arrange gtable[layout]
## 3 3 (2-4,1-2) arrange gtable[layout]
## 4 4 (3-4,3-4) arrange gtable[layout]Create an age model with Bacon
K <- runBacon(K,
lab.id.var = 'labID',
age.14c.var = 'age14C',
age.14c.uncertainty.var = 'age14CUncertainty',
age.var = 'age',
age.uncertainty.var = 'ageUncertainty',
depth.var = 'depth',
reservoir.age.14c.var = NULL,
reservoir.age.14c.uncertainty.var = NULL,
rejected.ages.var = NULL,
bacon.acc.mean = 10,
bacon.thick = 7,
accept.suggestions = TRUE,
ask = FALSE,
bacon.dir = "~/Cores",
suggest = FALSE,
close.connection = FALSE)## Using a mix of cal BP and calibrated C-14 dates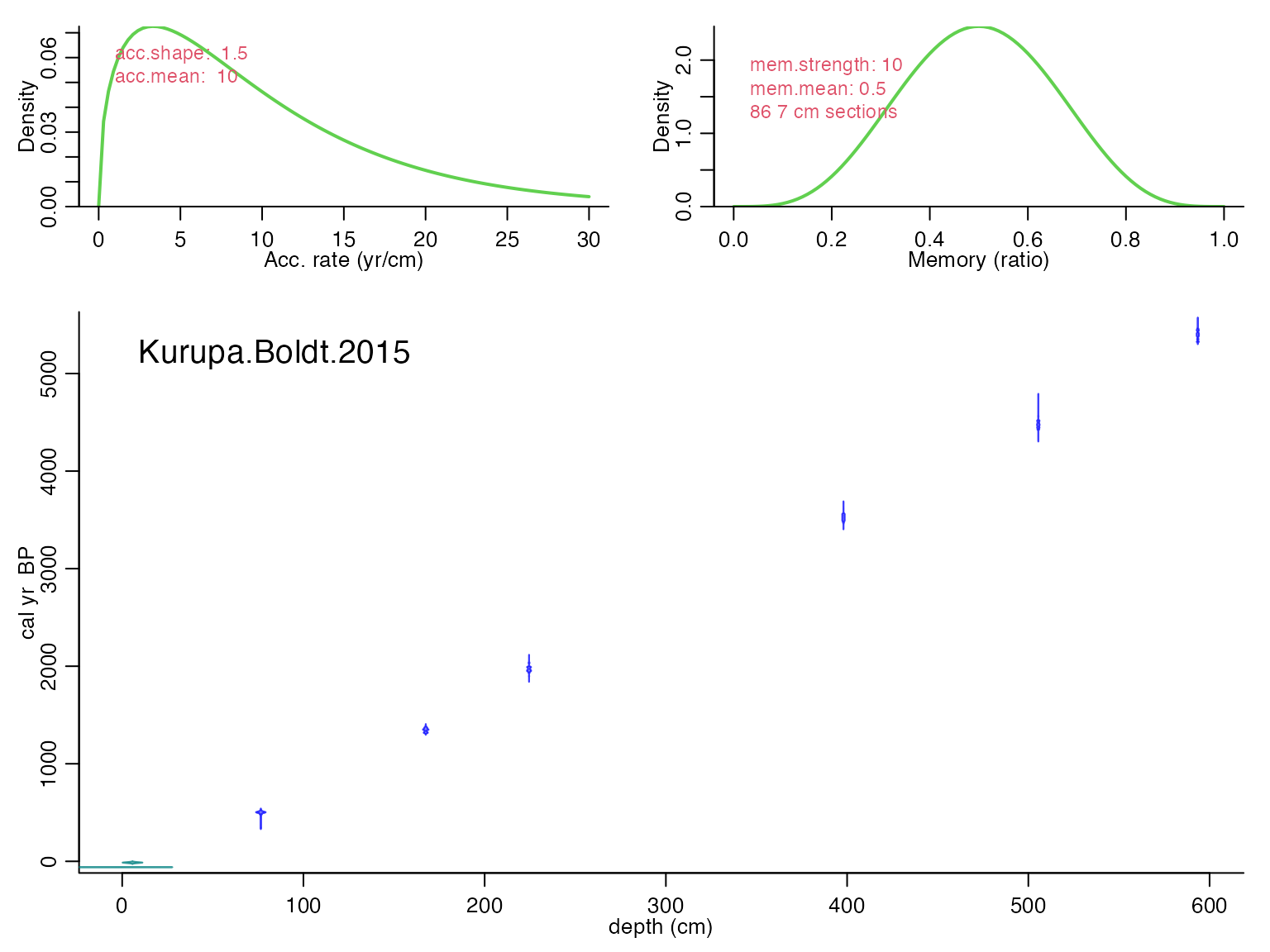
## Will run 16,500,000 iterations and store 4,000## Good MCMC mixing (effective sample size=541.42, >200)## No sign of MCMC drift (z=0.04, <1.96), OK## Warning, this will take quite some time to calculate. I suggest increasing d.by to, e.g., 10## Calculating age ranges...## 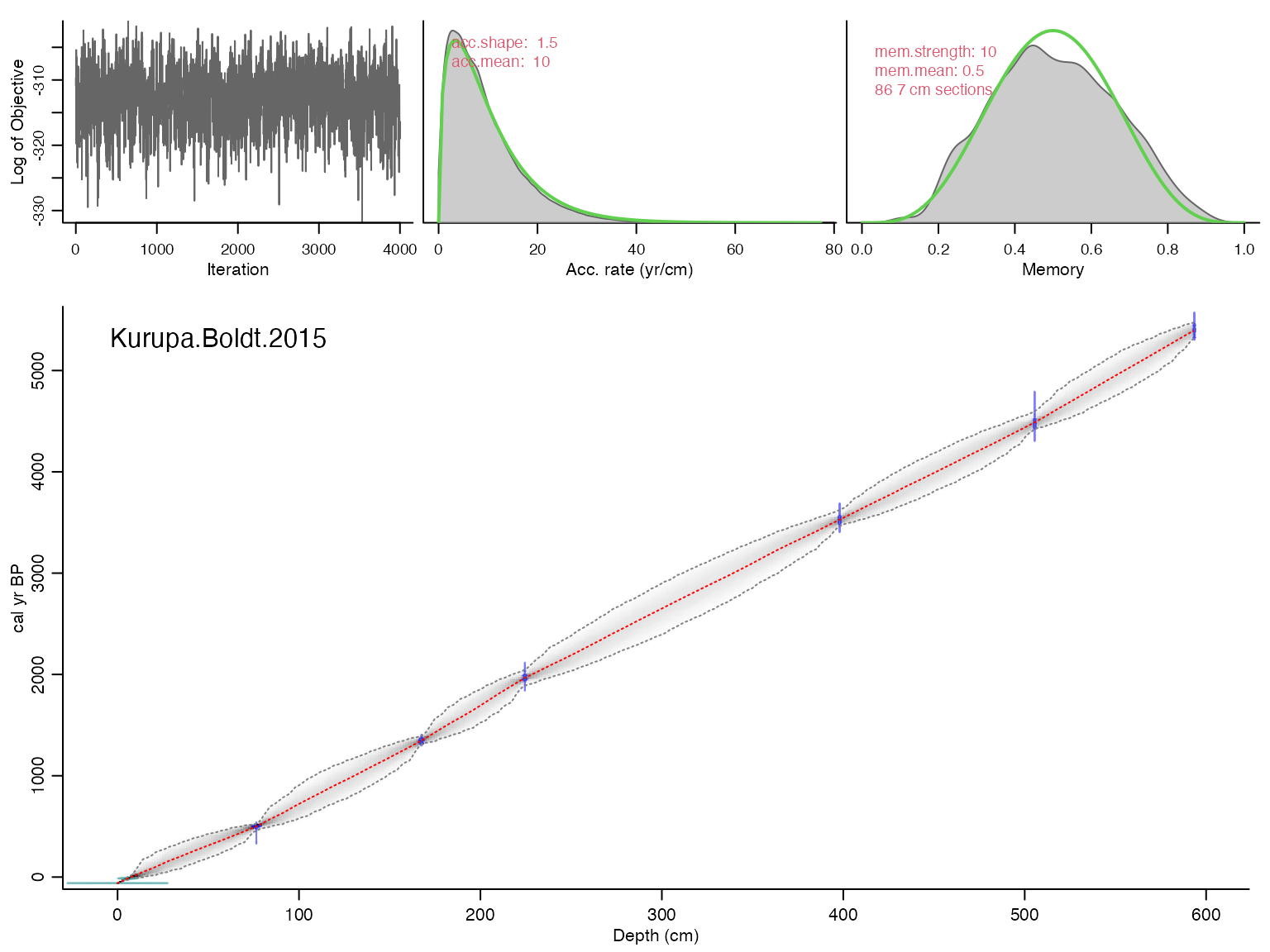
## Preparing ghost graph...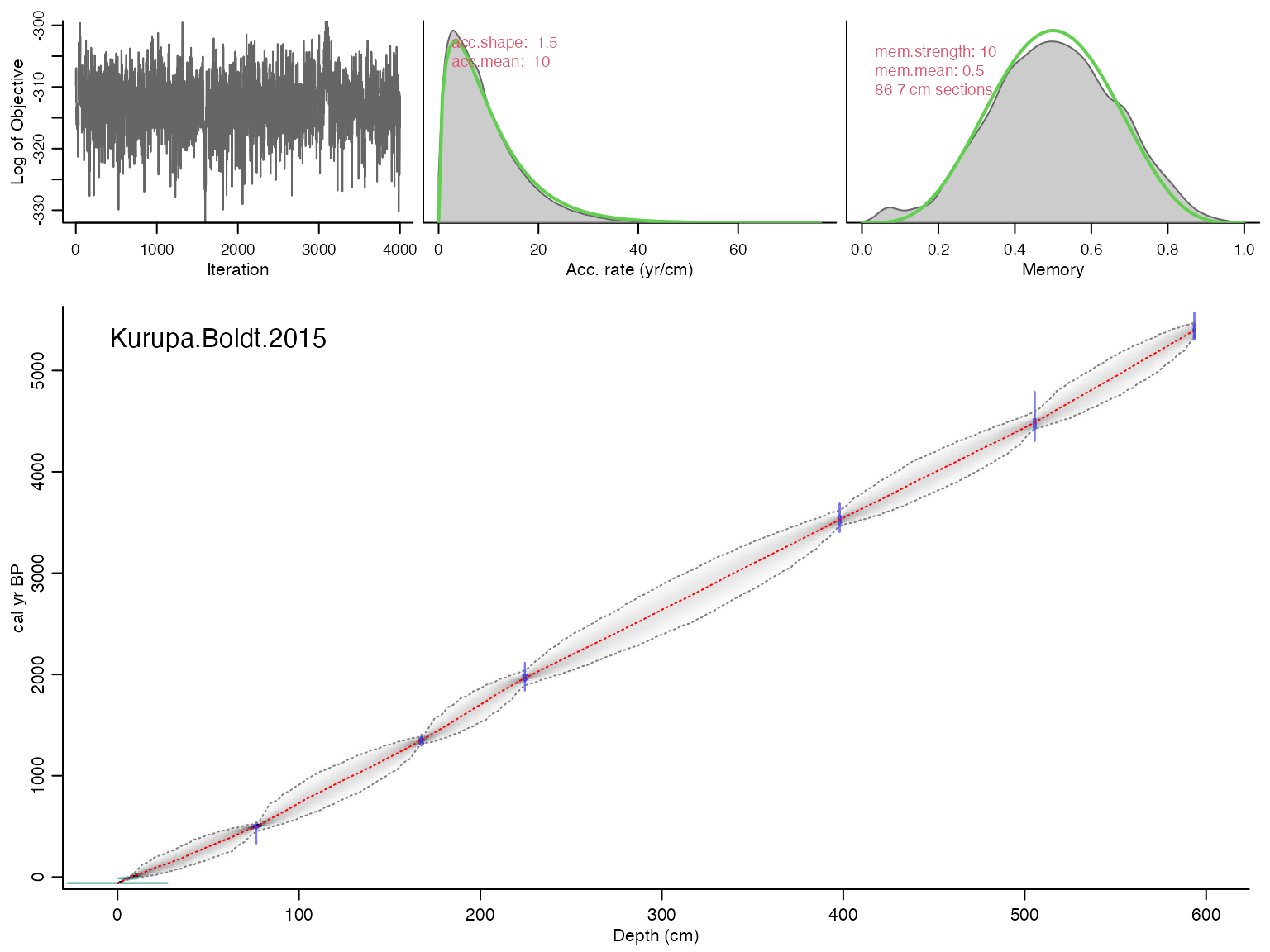
##
## Mean 95% confidence ranges 319 yr, min. 4 yr at 0 cm, max. 521 yr at 322 cm## 100% of the dates overlap with the age-depth model (95% ranges)## Posteriors: accrate mean 9.18, shape 1.59, memory mean 0.5, strength 8.34## Good MCMC mixing (effective sample size=541.42, >200)## No sign of MCMC drift (z=0.04, <1.96), OK## Warning, this will take quite some time to calculate. I suggest increasing d.by to, e.g., 10## And plot the ensemble output
plotChron(K,age.var = "ageEnsemble",dist.scale = 0.2)## [1] "Found it! Moving on..."
## [1] "Found it! Moving on..."
## [1] "plotting your chron ensemble. This make take a few seconds..."## Scale for x is already present.
## Adding another scale for x, which will replace the existing scale.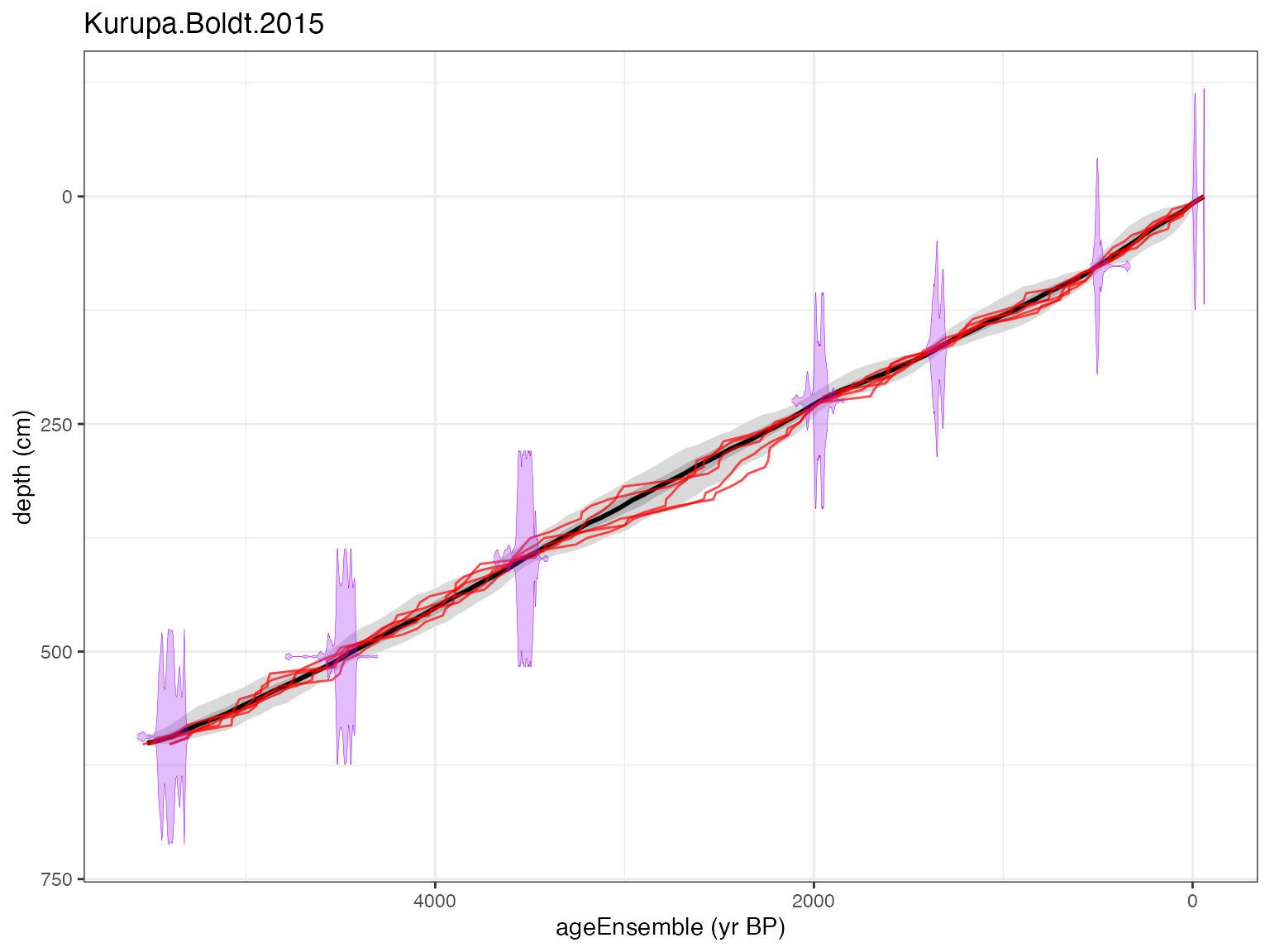
Map the age ensemble to the paleodata table
This is to get ensemble age estimates for each depth in the paleoData measurement table
K <- mapAgeEnsembleToPaleoData(K,age.var = "ageEnsemble")## [1] "Kurupa.Boldt.2015"
## [1] "Looking for age ensemble...."
## [1] "Found it! Moving on..."
## [1] "Found it! Moving on..."
## [1] "getting depth from the paleodata table..."
## [1] "Found it! Moving on..."
## mapAgeEnsembleToPaleoData created new variable ageEnsemble in paleo 1 measurement table 1
## mapAgeEnsembleToPaleoData also created new variable ageMedian in paleo 1 measurement table 1Select the paleodata age ensemble, and RABD data that we’d like to regress and calibrate
kae <- selectData(K,"ageEnsemble")## [1] "Found it! Moving on..."
rabd <- selectData(K,"RABD")## [1] "Found it! Moving on..."Now load in the instrumental data we want to correlate and regress agains
kurupa.instrumental <- readr::read_csv("http://lipdverse.org/geoChronR-examples/KurupaInstrumental.csv")## Rows: 134 Columns: 2
## ── Column specification ────────────────────────────────────────────────────────
## Delimiter: ","
## dbl (2): Year (AD), JJAS Temperature (deg C)
##
## ℹ Use `spec()` to retrieve the full column specification for this data.
## ℹ Specify the column types or set `show_col_types = FALSE` to quiet this message.Check age/time units before proceeding
kae$units## [1] "yr BP"yep, we need to convert the units from BP to AD
kae <- convertBP2AD(kae)And plot the output
Note that here we use the “Effective-N” significance option as we mimic the Boldt et al. (2015) paper.
plotCorEns(corout,significance.option = "eff-n")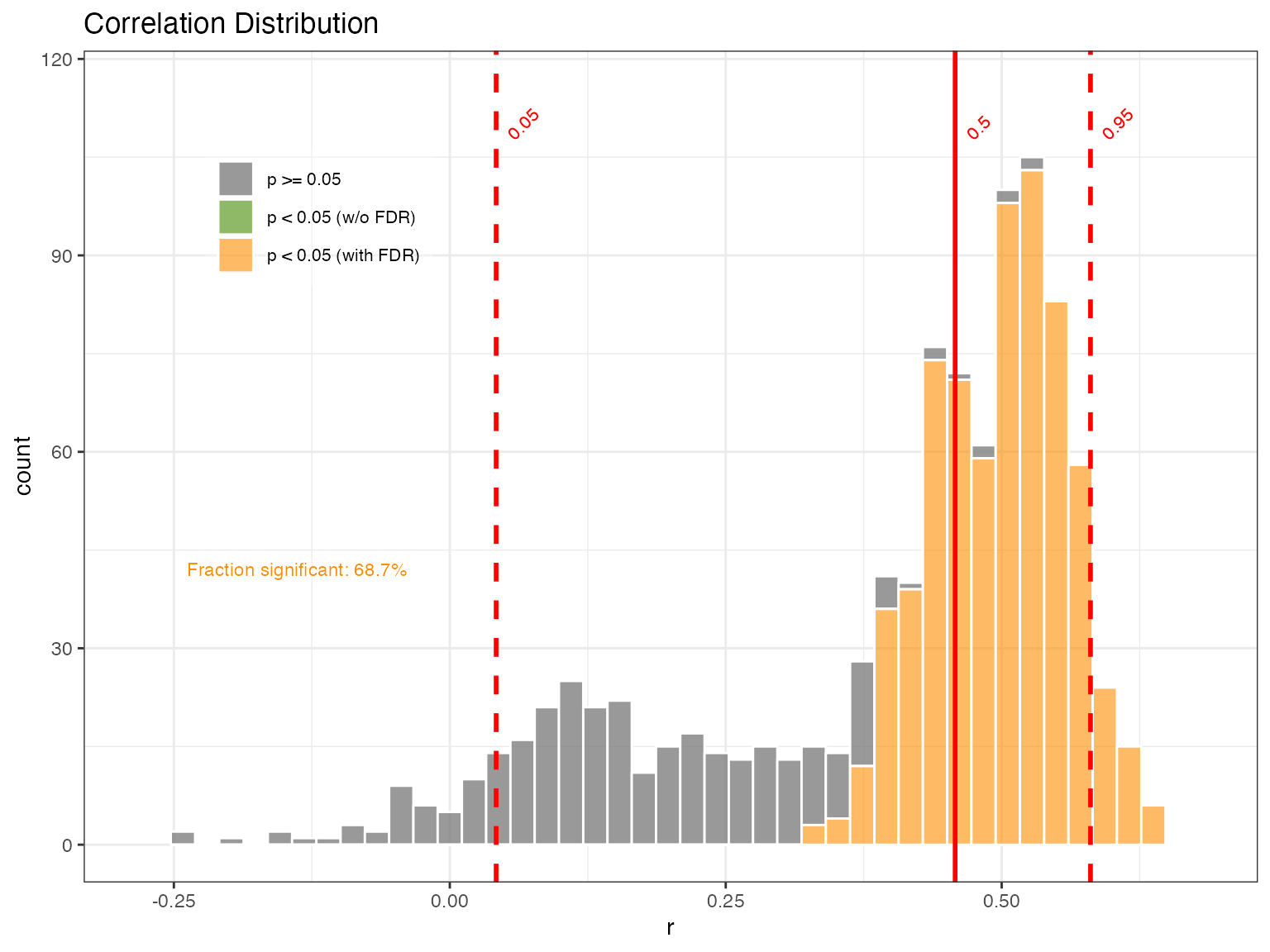
Mixed results. But encouraging enough to move forward.
Perform ensemble regression
OK, you’ve convinced yourself that you want to use RABD to model temperature back through time. We can do this simply (perhaps naively) with regession, and lets do it with age uncertainty, both in the building of the model, and the reconstructing
regout <- regressEns(time.x = kae,
values.x = rabd,
time.y =kyear,
values.y =kinst,
bin.step=3,
gaussianize = FALSE,
recon.bin.vec = seq(-4010,2010,by=20))And plot the output
regPlots <- plotRegressEns(regout,alp = 0.01,font.size = 8)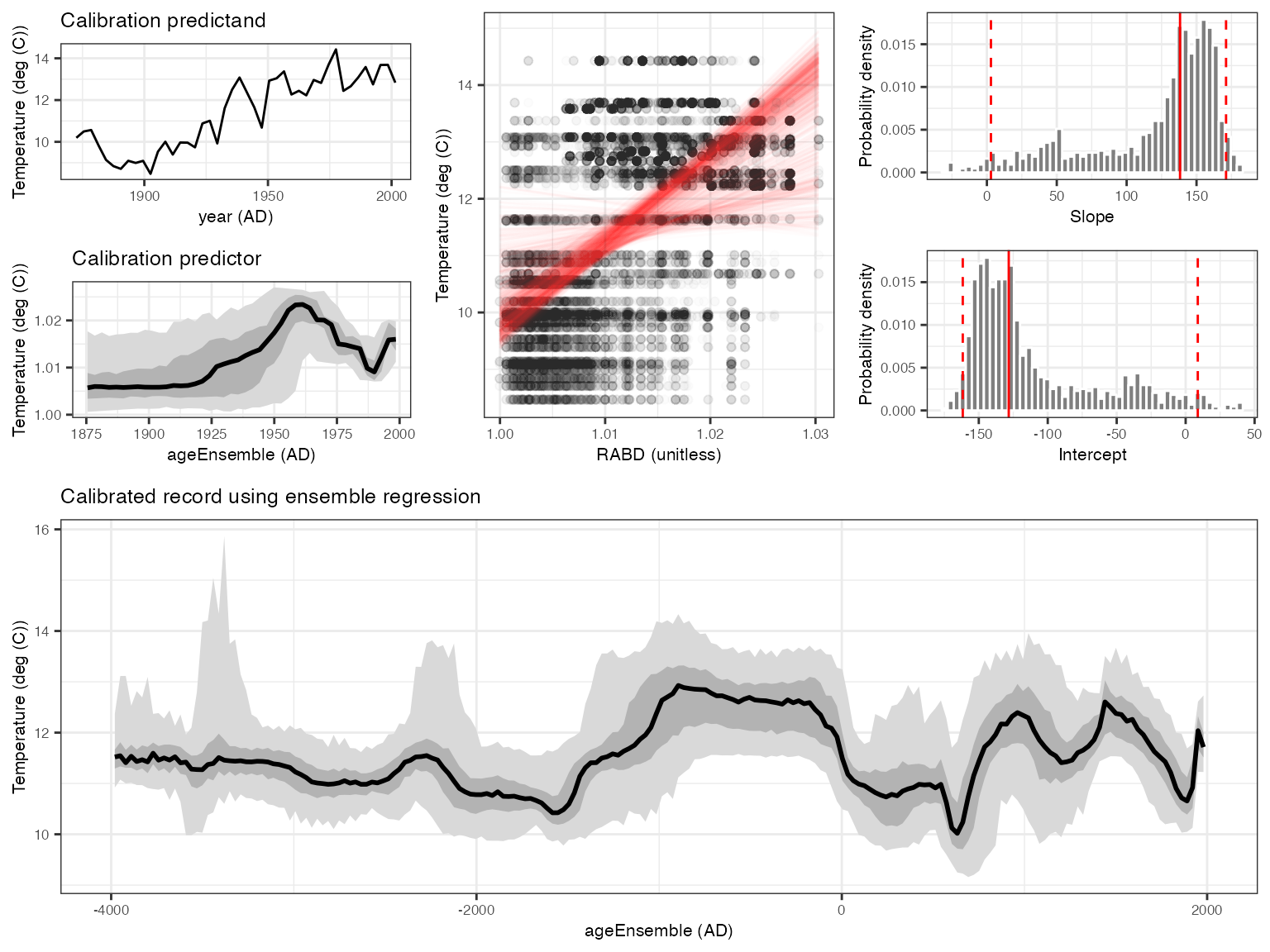
This result is consistent with that produced by Boldt et al., (2015), and was much simpler to produce with geoChronR.
In the next vignette learn about spectral analysis in geoChronR.
```