Tidy filtering, mapping and plotting in iso2k
Nick McKay
2025-05-31
Source:vignettes/tidyIso2k.Rmd
tidyIso2k.RmdLiPD filtering and mapping
This vignette highlights how to filter, plot and map a library of LiPD data, but this time using the tidyverse framework. As before first let’s load version 1.0.0 of the iso2k database. Check out the details of the iso2k dataset Earth System Science Data.
iso <- readLipd("http://lipdverse.org/iso2k/1_0_0/iso2k1_0_0.zip")That was easy - note that readLipd() can take individual LiPD files (ending in .lpd), or zip files full of LiPD files, directly from the web.
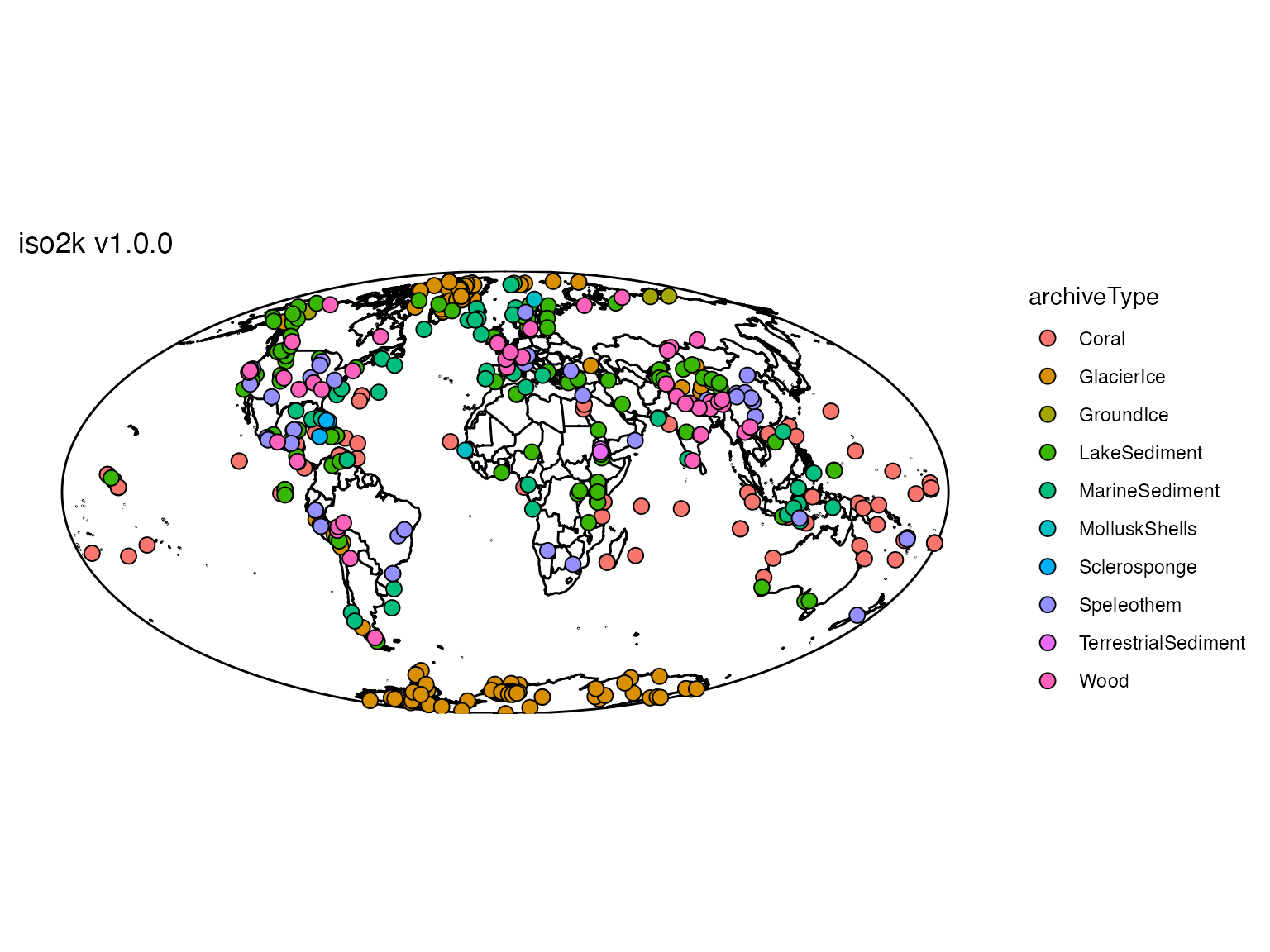
Now that the data are loaded, we can quickly map a bunch of LiPD
files using mapLipd().
Now we’ve seen an overview of all the files we’ve loaded, but now we’d like to filter the data.
First, we’ll extract a TS object, the primary way to work with multiple LiPD files. A TS is a list object that has an entry for each, column, and is readily queryable.
TS <- extractTs(iso)If you’re used to working in the tidyverse paradigm, you can also convert the TS object into a “tidyTs”, which is a large, long, tidy, tibble.
tTS <- tidyTs(TS,age.var = "year")Now we’ll filter it based on a range of criteria.
iTS <- filter(tTS, paleoData_iso2kPrimaryTimeseries == TRUE,
between(geo_latitude,66.5,90),
between(year,1000,2000),
paleoData_variableName == "d18O") %>%
arrange(geo_latitude)OK, now we’ve filtered down to just Arctic
data during the past Millennium. We can use mapTs() to map
the data coverage represented in iTS.
tm <- mapTs(iTS,
projection = "stereographic",#for polar projections
global = FALSE,
color = "archiveType", #color by the archiveType
shape = "paleoData_inferredMaterialGroup", #shape by the inferredMaterialGroup
size = 4,
bound.circ = TRUE
)
print(tm)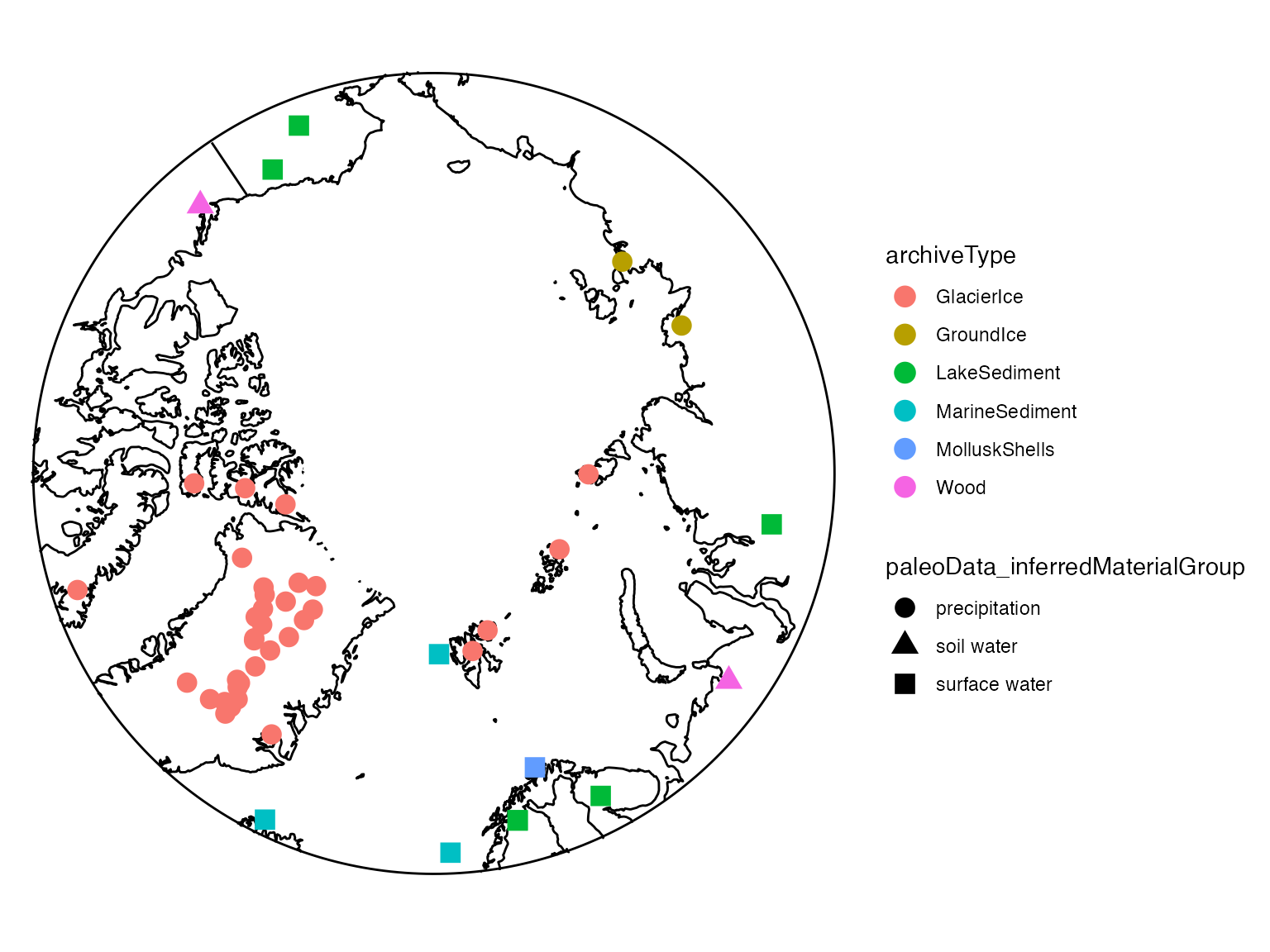
You can change the mapping options by passing options to
basemap() which creates the base map. Try
?basemap for options.
Time availability
You can also quickly make a plot of availability through time.
plotTimeAvailabilityTs(iTS,age.range = c(1000,2000),age.var = "year",step = 20)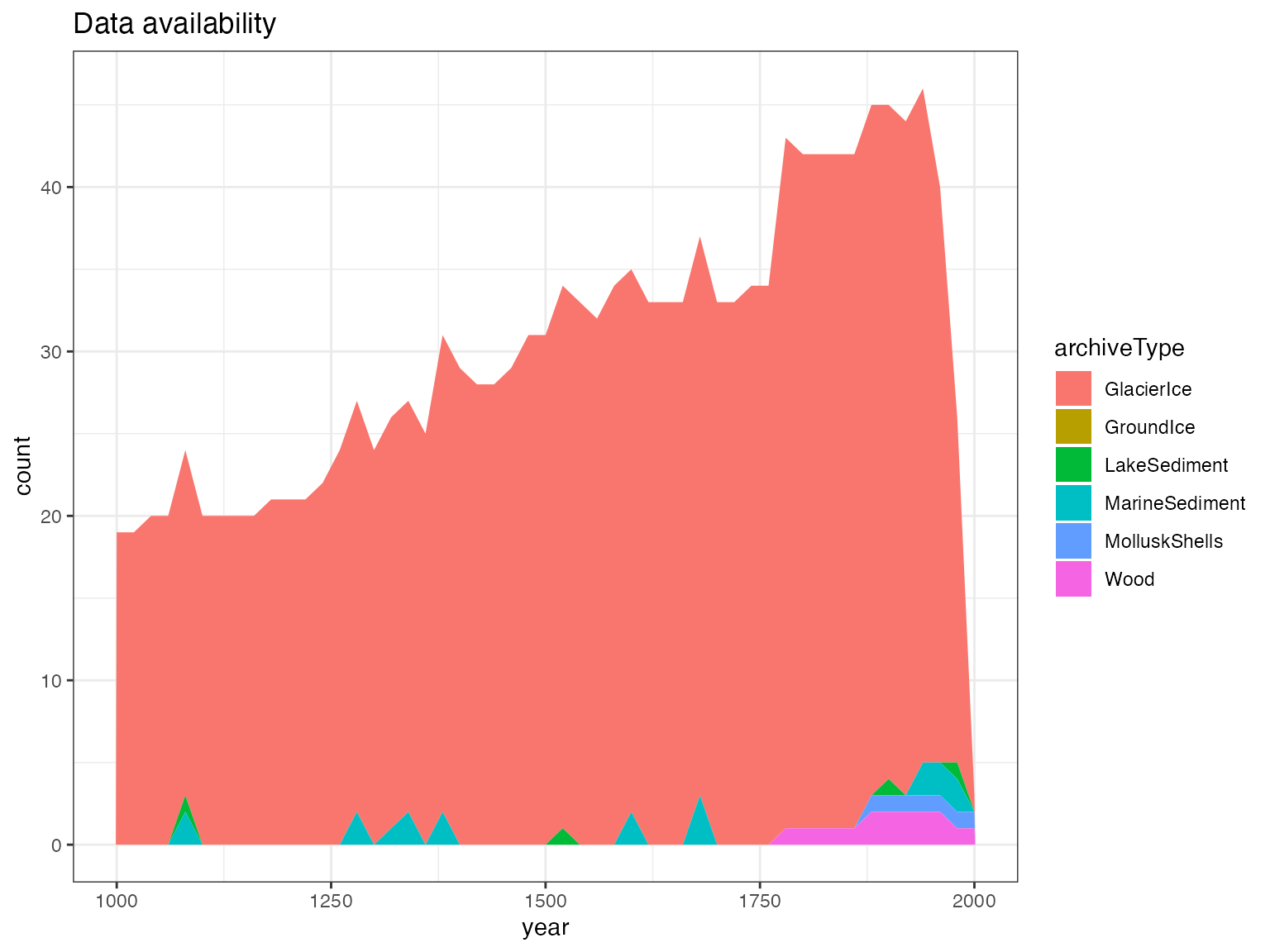
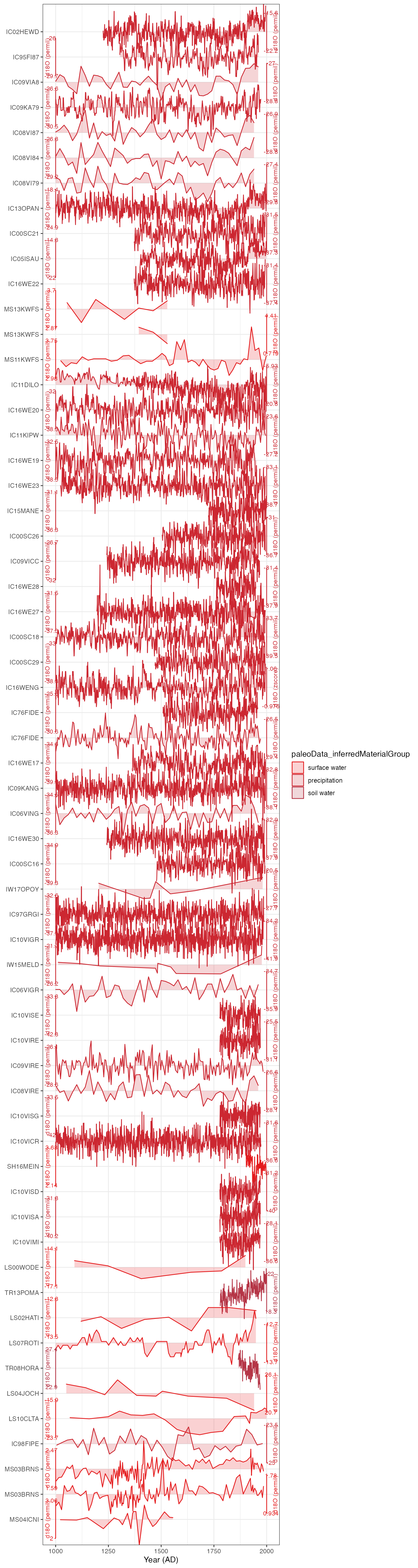
Plot a stack
Finally, because we’re working with a “tidy” TS (a tibble), we can
plot all the data using plotTimeseriesStack(). See the vignette on
plotTimeseriesStack() for more details of how to use this function.
Here we’ll plot all of these Arctic
datasets for the past millennium.
plotTimeseriesStack(iTS,color.var = "paleoData_inferredMaterialGroup",
color.ramp = function(nColors){RColorBrewer::brewer.pal(nColors,"Set1")})